BioMicroCenter:Oxford Nanopore Technologies: Difference between revisions
No edit summary |
|||
| Line 1: | Line 1: | ||
{{BioMicroCenter}} | {{BioMicroCenter}} | ||
<br><br> | <br><br> | ||
The MIT BioMicro Center has | The MIT BioMicro Center has two PromethION P2 Solos. We support de novo sequencing and have tested some RNA-seq metrics on these platforms. Each flowcell can potentially accommodate many barcoded samples (depending on sample quality and desired coverage) and each unit can theoretically run up to 2 flowcells simultaneously depending on the projects. | ||
<br><br> | <br><br> | ||
==Oxford Nanopore Sequencing== | ==Oxford Nanopore Sequencing== | ||
Revision as of 13:33, 25 April 2025
HOME -- SEQUENCING -- LIBRARY PREP -- HIGH-THROUGHPUT -- COMPUTING -- OTHER TECHNOLOGY
The MIT BioMicro Center has two PromethION P2 Solos. We support de novo sequencing and have tested some RNA-seq metrics on these platforms. Each flowcell can potentially accommodate many barcoded samples (depending on sample quality and desired coverage) and each unit can theoretically run up to 2 flowcells simultaneously depending on the projects.
Oxford Nanopore Sequencing
|
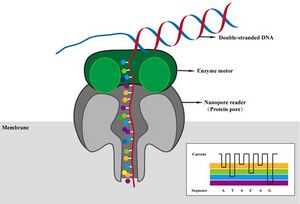
A variety of preparation kits allow DNA, cDNA, or direct RNA to be sequenced. The input amount necessary for each type of sample varies according to the desired result. For the longest reads of genomic DNA, micrograms worth of clean starting material provide the highest quality of data, but not the highest amounts of reads. For amplicons including cDNA made from RNA, amplifying hundreds of nanograms should be simple, thus a lower input in the picograms should be reasonable.
|
Oxford Nanopore Sequencers
| SPEC | PromethiON P2 Solo |
|---|---|
| SEQUENCER |  |
| AVG READS/FLOWCELL |
50 Gb |
| MAX TIME/FLOWCELL |
3 days (adjustable) |
| BEST FOR |
|