BioMicroCenter:Singular Sequencing: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
== Singular Sequencing == | == Singular Sequencing == | ||
The Center currently hosts a G4 platform by [https://singulargenomics.com/| Singular Genomics]. Singular’s G4 platform is a short-read mid-throughput sequencing platform delivering adequate sequencing data through its proprietary sequencing-by-synthesis chemistry at a significantly competitive price point. | The Center currently hosts a G4 platform by [https://singulargenomics.com/| Singular Genomics]. Singular’s G4 platform is a short-read mid-throughput sequencing platform delivering adequate sequencing data through its proprietary sequencing-by-synthesis chemistry at a significantly competitive price point. | ||
{| class="wikitable" border=1 | {| | ||
!width= | | valign="top" | | ||
!width= | {| class="wikitable" border=1 | ||
!width=150| Service | |||
!width=300| Singular Sequencing | |||
|- | |- | ||
|INPUT || Singular libraries | |INPUT || Singular libraries | ||
| Line 40: | Line 42: | ||
|- | |- | ||
|} | |} | ||
| valign="top" | | |||
=== ANCHORS === | |||
Singular requires S1/S2 anchors in place of Illumina P5/P7 anchors. These can be replaced 1:1 in most oligo designs: | |||
*s1:ACAAAGGCAGCCACGCACTCCTTCCCTGT | |||
*s2:CTCCAGCGAGATGACCCTCACCAACCACT | |||
=== | === INDEXING === | ||
Singular indexes are read from the Anchor sequences in all cases and custom primers for indexes are not allowed. <BR> | |||
Indexes are read from the S1 primer first (index1) followed by S2 (index2) <BR> | |||
Index reads are the SAME SEQUENCE as ordered in most classical library preps. <BR> | |||
Indexes are REQUIRED and should be 8 nucleotides in all lane by lane sequencing. | |||
*Min custom primer submission: 20 uL at 100 uM | |||
===LANE BY LANE=== | |||
*Must be 50 or 150 paired-end dual indexed | *Must be 50 or 150 paired-end dual indexed | ||
*Pool must be compatible with TruSeq, Nextera, TruSeq, smRNA and Solexa sequencing primers | *Pool must be compatible with TruSeq, Nextera, TruSeq, smRNA and Solexa sequencing primers | ||
=== MINIMUM READS === | |||
*the BMC has performed quality control | *the BMC has performed quality control. | ||
* | * No custom primers. | ||
*submitted libraries are at least 2 nM. | *submitted libraries are at least 2 nM. | ||
All other requests, including use of custom sequencing primers require a full flowcell. Illumina libraries which have been converted in the Center will also require a full flowcell. | All other requests, including use of custom sequencing primers require a full flowcell. Illumina libraries which have been converted in the Center will also require a full flowcell. | ||
|} | |||
{| | |||
| | |||
== The G4 Platform == | == The G4 Platform == | ||
{| class="wikitable" border=1 | {| class="wikitable" border=1 | ||
| Line 80: | Line 90: | ||
* 100nt | * 100nt | ||
* 300nt | * 300nt | ||
|- | |- | ||
|'''KEY NOTES'''|| | |'''KEY NOTES'''|| | ||
| Line 93: | Line 96: | ||
* 4-lanes per flow cell | * 4-lanes per flow cell | ||
* 4-flow cells per run | * 4-flow cells per run | ||
* | * Anchor replacement or anchor expansion of Illumina libraries | ||
|- | |- | ||
|'''THANKS TO'''|| | |'''THANKS TO'''|| | ||
* Singular Genomics | * Singular Genomics | ||
|} | |||
| valign='top' | | |||
{| | |||
! INDEX HOPING | |||
! ACCURACY | |||
|- | |||
| | |||
[[IMAGE:IndexhopF3.jpeg|left|250px|Index hopping for a standard set of Singular libraries]] | |||
| | |||
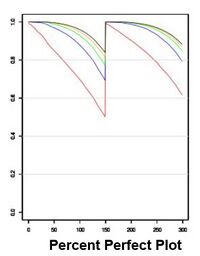
[[IMAGE:PPPPF3.jpeg|right|200px|Percent Perfect Plot for Singular libraries 150PE]] | |||
|- | |||
| Index hopping on Singular runs is on par with GAIIx and HiSeq. High levels of index hopping as observed on Illumina X-AMP chemistry is not observed. | |||
| Percent perfect plot for phiX on current Singular chemistry ([https://pmc.ncbi.nlm.nih.gov/articles/pmid/27672352/ Manley et al., 2016]) | |||
|} | |||
|} | |} | ||
Revision as of 16:12, 20 July 2025
Singular Sequencing
The Center currently hosts a G4 platform by Singular Genomics. Singular’s G4 platform is a short-read mid-throughput sequencing platform delivering adequate sequencing data through its proprietary sequencing-by-synthesis chemistry at a significantly competitive price point.
|
ANCHORSSingular requires S1/S2 anchors in place of Illumina P5/P7 anchors. These can be replaced 1:1 in most oligo designs:
INDEXINGSingular indexes are read from the Anchor sequences in all cases and custom primers for indexes are not allowed.
LANE BY LANE
MINIMUM READS
All other requests, including use of custom sequencing primers require a full flowcell. Illumina libraries which have been converted in the Center will also require a full flowcell.
|
The G4 Platform |
|