BioMicroCenter:SingleCell: Difference between revisions
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
[[File:SingleCellOptions.png|thumb|right|400px]] | [[File:SingleCellOptions.png|thumb|right|400px]] | ||
The BioMicro Center supports a broad range of methods for single cell sequencing. The choice of method depends heavily on the type of question being asked and the source material. The methods break down into those supporting | The BioMicro Center supports a broad range of methods for single cell sequencing. The choice of method depends heavily on the type of question being asked and the source material. The methods break down into those supporting individual cells characterized in single wells, methods that use droplet isolation and library preparation, and those that support rapid characterization of a modest number of transcripts using in situ sequencing (coming soon). These cell requirements for each method vary significantly so reviewing each method is valuable and we strongly encourage consultation with BMC staff prior to beginning the experiment. | ||
<BR><BR><BR><BR><BR><BR>. | <BR><BR><BR><BR><BR><BR>. | ||
== 10x CHROMIUM X == | == 10x CHROMIUM X == | ||
[[File:25_10xAssays.png|thumb|left|400px]] | |||
The BioMicro Center provides access to 10x Genomics library preparation as an assisted or a walk-up service. Added in 2023, The 10x Chromium X can handle a broad variety of methodologies now including [https://www.10xgenomics.com/solutions/single-cell/ 3', 5' and fixed RNA sequencing], [https://www.10xgenomics.com/solutions/single-cell-atac/ ATAC and multiome] <!-- [https://www.10xgenomics.com/solutions/single-cell-cnv/ CNV]. --> Dropoff for assisted service is coordinated with Center staff with at least one week's lead time. Users should bring their single cell/nuclei suspension(s) in 1.5 mL microfuge tubes in the standard buffer at or around that time. Staff works with the user to intake the initial samples and proceed through the protocol with quality control checks at the appropriate steps. | |||
<BR><BR> | |||
Chromium X usage is also offered as a walkup service. Usage may be scheduled on the iLabs calendar after training by BMC staff. This still requires at least a week's lead time before usage. You will not have permission to schedule the equipment until you have been approved by BMC staff. Reagents are stocked in the BioMicro Center and we ask that you use our reagents. This allows us to get larger bulk discounts we can pass on to our users. Please note that we do include a fraction of the instrument usage cost in the cost of the chips and will charge this cost if you use your own chips. | |||
<BR><BR> | |||
The full 10x suite of software is installed on LURIA and is integrated into our analysis package. <BR><BR> | |||
{| | {| | ||
|- style="vertical-align: top;" | |- style="vertical-align: top;" | ||
| Line 96: | Line 102: | ||
=== 10X Genomics === | === 10X Genomics === | ||
[[image:10xX.jpg|thumb|right|500px|10x Chromium X]] | [[image:10xX.jpg|thumb|right|500px|10x Chromium X]] | ||
FAQs for Users <BR> | FAQs for Users <BR> | ||
Revision as of 13:15, 2 September 2025
HOME -- SEQUENCING -- LIBRARY PREP -- HIGH-THROUGHPUT -- COMPUTING -- OTHER TECHNOLOGY
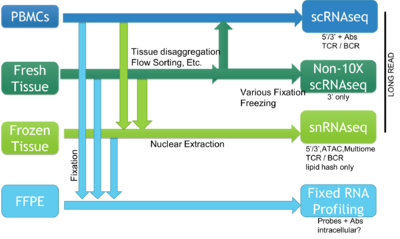
The BioMicro Center supports a broad range of methods for single cell sequencing. The choice of method depends heavily on the type of question being asked and the source material. The methods break down into those supporting individual cells characterized in single wells, methods that use droplet isolation and library preparation, and those that support rapid characterization of a modest number of transcripts using in situ sequencing (coming soon). These cell requirements for each method vary significantly so reviewing each method is valuable and we strongly encourage consultation with BMC staff prior to beginning the experiment.
.
10x CHROMIUM X
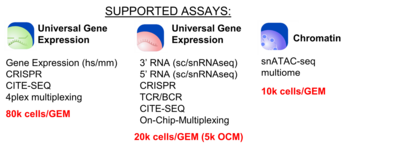
The BioMicro Center provides access to 10x Genomics library preparation as an assisted or a walk-up service. Added in 2023, The 10x Chromium X can handle a broad variety of methodologies now including 3', 5' and fixed RNA sequencing, ATAC and multiome Dropoff for assisted service is coordinated with Center staff with at least one week's lead time. Users should bring their single cell/nuclei suspension(s) in 1.5 mL microfuge tubes in the standard buffer at or around that time. Staff works with the user to intake the initial samples and proceed through the protocol with quality control checks at the appropriate steps.
Chromium X usage is also offered as a walkup service. Usage may be scheduled on the iLabs calendar after training by BMC staff. This still requires at least a week's lead time before usage. You will not have permission to schedule the equipment until you have been approved by BMC staff. Reagents are stocked in the BioMicro Center and we ask that you use our reagents. This allows us to get larger bulk discounts we can pass on to our users. Please note that we do include a fraction of the instrument usage cost in the cost of the chips and will charge this cost if you use your own chips.
The full 10x suite of software is installed on LURIA and is integrated into our analysis package.
scRNAseq / snRNAseq
CHROMATIN
|
10X Genomics
2) How many cells are captured in the Assay?
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
NAMOCELL SINGLE CELL SORTER
|
The Namocell Single Cell Sorter allows users to sort cells into plates. The sorter uses microfluidics to sort single cells in 1uL of sheath fluid into a well. The instrument uses disposable cartridges to minimize contamination. The instrument integrates well with the TTP Labtech Mosquito HV which handles small reaction volumes.  |

