BioMicroCenter:SingleCell: Difference between revisions
No edit summary |
|||
| (33 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{BioMicroCenter}} | {{BioMicroCenter}} | ||
The BioMicro Center supports | [[File:SingleCellOptions.png|thumb|right|400px]] | ||
The BioMicro Center supports a broad range of methods for single cell sequencing. The choice of method depends heavily on the type of question being asked and the source material. The methods break down into those supporting individual cells characterized in single wells, methods that use droplet isolation and library preparation, and those that support rapid characterization of a modest number of transcripts using in situ sequencing (coming soon). These cell requirements for each method vary significantly so reviewing each method is valuable and we strongly encourage consultation with BMC staff prior to beginning the experiment. | |||
<BR><BR><BR><BR><BR><BR>. | |||
<!-- | |||
== 10x CHROMIUM X == | |||
[[File:25_10xAssays.png|thumb|left|400px]] | |||
The BioMicro Center provides access to 10x Genomics library preparation as an assisted or a walk-up service. Added in 2023, The 10x Chromium X can handle a broad variety of methodologies now including [https://www.10xgenomics.com/solutions/single-cell/ 3', 5' and fixed RNA sequencing], [https://www.10xgenomics.com/solutions/single-cell-atac/ ATAC and multiome] <!-- [https://www.10xgenomics.com/solutions/single-cell-cnv/ CNV]. --> Dropoff for assisted service is coordinated with Center staff with at least one week's lead time. Users should bring their single cell/nuclei suspension(s) in 1.5 mL microfuge tubes in the standard buffer at or around that time. Staff works with the user to intake the initial samples and proceed through the protocol with quality control checks at the appropriate steps. | |||
<BR><BR> | |||
Chromium X usage is also offered as a walkup service. Usage may be scheduled on the iLabs calendar after training by BMC staff. This still requires at least a week's lead time before usage. You will not have permission to schedule the equipment until you have been approved by BMC staff. Reagents are stocked in the BioMicro Center and we ask that you use our reagents. This allows us to get larger bulk discounts we can pass on to our users. Please note that we do include a fraction of the instrument usage cost in the cost of the chips and will charge this cost if you use your own chips. | |||
<BR><BR> | |||
The full 10x suite of software is installed on LURIA and is integrated into our analysis package. <BR><BR> | |||
{| | {| | ||
|- style="vertical-align: top;" | |- style="vertical-align: top;" | ||
|style="width: | |style="width: 450px;"| | ||
=== scRNAseq / snRNAseq === | |||
{| class="wikitable" border=1 | {| class="wikitable" border=1 | ||
! | ! | ||
! | ! 3'RNA/5'RNA <BR> (GEM-X) | ||
! 3'/5' OnChipMultiplexing <BR> (OCM) | |||
! Fixed RNA <BR> (Flex GEM-X) | |||
|- | |- | ||
| | !INPUT | ||
|colspan="2"| | |||
[https://www.10xgenomics.com/support/universal-three-prime-gene-expression/documentation/steps/sample-prep Freshly counted cells/nuclei in suspension.] | |||
| | | | ||
[https://www.10xgenomics.com/support/flex-gene-expression/documentation/steps/sample-prep Fixed cells in suspension.] | |||
|- | |- | ||
| | !Cell Concentration <BR><small> cells|nuclei/uL </small> | ||
| | |100-2000 | ||
|100-2000 | |||
|2000+ | |||
|- | |- | ||
| | ! Fraction Recovered <BR><small> est. percent of input cells captured </small> | ||
| ~65% | |||
| ~60% | |||
| ~50% | |||
|- | |- | ||
| | ! Target recovery <BR> <small> multiply by recovery for input </small> | ||
| 500-20,000/lane | |||
| 500-5,000/lane | |||
| 500-10,000/sample | |||
|- | |- | ||
| | ! Expected doublet rate @ Max load | ||
| 8% | |||
| 7.5% | |||
| 8% doublet @ 10k | |||
|- | |- | ||
| | !Multiplexing | ||
| 1 sample | |||
| 4 samples/library | |||
| 4 samples/lane | |||
|- | |- | ||
| | !ADD'L AMPLICONS | ||
|colspan="2" | | |||
Custom priming for CITEseq, CRISPRs (perturb-seq), TCR (5'), BCR (5'). <BR> Please provide custom oligos and protocols to core. | |||
| | |||
Additional genes [https://www.10xgenomics.com/support/flex-gene-expression/documentation/steps/experimental-design-and-planning/custom-probe-design-for-visium-spatial-gene-expression-and-chromium-single-cell-gene-expression-flex require new probes.] | |||
-- | |||
|- | |- | ||
! | ! KEY NOTES | ||
| | | | ||
| | |||
| Human/Mouse only | |||
| | |||
| | |||
|- | |- | ||
!SUBMISSION | !SUBMISSION FORMS | ||
|colspan="3"| ASSISTED - [https://mit.ilabsolutions.com/service_item/new/3381?spt_id=12903 ilabs] | |colspan="3"| ASSISTED - [https://mit.ilabsolutions.com/service_item/new/3381?spt_id=12903 ilabs] WALKUP - [https://mit.ilabsolutions.com/equipment/380963 Calendar] | ||
|- | |- | ||
!DELIVERY | !DELIVERY | ||
|colspan="3"| FASTQ, SAM, BAM, 10X QC, loupe file | |colspan="3"| FASTQ, SAM, BAM, 10X QC, loupe file | ||
|- | |- | ||
!DONATED BY | !DONATED BY | ||
|colspan="3"| [http://mit.edu/manoli/ Prof. Manolis Kellis] | |colspan="3"| [http://mit.edu/manoli/ Prof. Manolis Kellis] | ||
|} | |||
=== CHROMATIN === | |||
{| class="wikitable" border=1 | |||
! | |||
! snATACseq | |||
! multiome | |||
|- | |||
! Input | |||
| colspan=2 | | |||
* 160-8,000 nuclei/uL | |||
* 50% estimated recovery | |||
* 7.5% doublets @ 10k cells recovered | |||
* 10uL minimum volume | |||
|- | |||
! Multiplexing | |||
| colspan =2 | Not available | |||
|- | |||
!SUBMISSION FORMS | |||
|colspan="2"| ASSISTED - [https://mit.ilabsolutions.com/service_item/new/3381?spt_id=12903 ilabs] WALKUP - [https://mit.ilabsolutions.com/equipment/380963 Calendar] | |||
|- | |- | ||
! | !DELIVERY | ||
|colspan=" | |colspan="2"| FASTQ, BAM, 10X QC, loupe file | ||
|} | |} | ||
| | |||
=== 10X Genomics === | |||
[[image:10xX.jpg|thumb|right|500px|10x Chromium X]] | [[image:10xX.jpg|thumb|right|500px|10x Chromium X]] | ||
FAQs for Users <BR> | FAQs for Users <BR> | ||
'''1) What buffers are compatible with 10X applications?''' | '''1) What buffers in the final suspension are compatible with 10X applications?''' | ||
<br> | <br> | ||
Buffers/media should not contain excessive amounts of EDTA (>0.1 mM) or magnesium (>3 mM) and should be free of surfactants (i.e. Tween-20, SDS etc) and any RNases or DNases. | [https://kb.10xgenomics.com/hc/en-us/articles/115001937123-What-buffers-can-be-used-for-washing-and-cell-resuspension Buffers/media] for the submitted single cell/nuclei suspension should not contain excessive amounts of EDTA (>0.1 mM) or magnesium (>3 mM) and should be free of surfactants (i.e. Tween-20, SDS etc) and any RNases or DNases. | ||
*1xPBS (calcium free and magnesium free) containing 0.04% weight/volume BSA (400 µg/ml) is recommended for most general protocols and is considered the standard buffer | *1xPBS (calcium free and magnesium free) containing 0.04% weight/volume BSA (400 µg/ml) is recommended for most general protocols and is considered the standard buffer | ||
*Cell culture media with up to 1% BSA or up to 10% FBS if cells are not viable in standard buffer | *Cell culture media with up to 1% BSA or up to 10% FBS if cells are not viable in standard buffer | ||
| Line 91: | Line 113: | ||
*Nuclei also require addition of RNase Inhibitor along with 10X Genomics 1X nuclei buffer before chip loading, instructions for which are included in 10x user guides. If needed, users can collect buffer aliquots from the BMC after submitting a project. Please coordinate with BMC staff for pick-up. <br> | *Nuclei also require addition of RNase Inhibitor along with 10X Genomics 1X nuclei buffer before chip loading, instructions for which are included in 10x user guides. If needed, users can collect buffer aliquots from the BMC after submitting a project. Please coordinate with BMC staff for pick-up. <br> | ||
'''2) How many cells are captured in the Assay?''' | '''2) How many cells are captured in the Assay?''' | ||
<br>Up to 10,000 cells for NextGEM and 20,000 cells for GEM-X can be uniquely barcoded, but this highly depends on cell counts and viability. Dying cells will leak RNA, hence may not be captured efficiently leading to sample failures. We recommend to count cells at the BMC to avoid discrepancies, but can work with users' counts as well. <BR> | |||
'''3) What are the best practices for flow sorting cells? | '''3) What are the best practices for flow sorting cells? <br> | ||
-https://kb.10xgenomics.com/hc/en-us/articles/360048826911-What-are-the-best-practices-for-flow-sorting-cells-for-10x-Genomics-assays < | 10x provides guidance with their tested protocols about pre-sort buffer, collection buffer and FACS best practices [https://kb.10xgenomics.com/hc/en-us/articles/360048826911-What-are-the-best-practices-for-flow-sorting-cells-for-10x-Genomics-assays here.] <br> | ||
'''4) What is the expected size distribution for cDNA?''' | '''4) What is the expected size distribution for cDNA?''' <br> | ||
cDNA for 3' and 5' libraries will span between 400 to 9000 base pairs, depending on sample type. | |||
<br> | |||
For more resources, please visit https://kb.10xgenomics.com/hc/en-us | '''5) How much sequencing per sample is recommended?''' <br> | ||
10x makes several recommendations in their [https://www.10xgenomics.com/support/epi-atac/documentation/steps/sequencing/sequencing-handbook sequencing handbook]. Recommendation numbers vary by sample type, expected CNPs per sample, assay type and general sample quality. The higher the CNPs, the higher the quality, the more likely increased read depth is required. <br> | |||
For more resources, please visit https://kb.10xgenomics.com/hc/en-us <br> | |||
| | | | ||
|} | |} | ||
| Line 136: | Line 160: | ||
|} | |} | ||
| | | | ||
[[image:Namo.jpg|thumb|right|Namo (Namocell) Bio-Techne]] | |||
The [https://www.namocell.com/namo/ Namocell Single Cell Sorter] allows users to sort cells into plates. The sorter uses microfluidics to sort single cells in 1uL of sheath fluid into a well. The instrument uses disposable cartridges to minimize contamination. The instrument integrates well with the [[BioMicroCenter:Tecan_Freedom_Evo#TTP_LABTECH_MOSQUITO_HV|TTP Labtech Mosquito HV]] which handles small reaction volumes. <BR><BR> | The [https://www.namocell.com/namo/ Namocell Single Cell Sorter] allows users to sort cells into plates. The sorter uses microfluidics to sort single cells in 1uL of sheath fluid into a well. The instrument uses disposable cartridges to minimize contamination. The instrument integrates well with the [[BioMicroCenter:Tecan_Freedom_Evo#TTP_LABTECH_MOSQUITO_HV|TTP Labtech Mosquito HV]] which handles small reaction volumes. <BR><BR> | ||
|} | |} | ||
<BR><BR>. | |||
<!-- | == AVITI24 IN SITU SEQUENCING == | ||
*** coming in early 2026 *** | |||
The AVITI24 enables direct in situ sequencing of samples, allowing for very rapid characterization of single cells. Cells can be grown or placed on the AVITI24 flowcell and processed. Runs are predicted to produce a few thousand reads plus cytostaining and antibody labeling, all within a day. More information will be released soon. | |||
<!-- commenting out Seq-Well | |||
== SEQ-WELL == | |||
{| | |||
|- style="vertical-align: top;" | |||
|style="width: 300px;"| | |||
{| class="wikitable" border=1 | |||
! | |||
!SEQ-WELL | |||
|- | |||
|INPUT | |||
| | |||
* FULL PREP: <BR>50,000 single free cells. 90% viability | |||
* EXO1 and AMP <BR> Tubes with cDNA attached to beads | |||
* LIBRARY ONLY <BR> Amplified cDNA | |||
|- | |||
|THROUGHPUT | |||
| | |||
* FULL PREP/EXO: 4 samples/day | |||
* LIBRARY: 24 samples/day | |||
|- | |||
|INCLUDED || QC at cDNA stage, library generation. | |||
|- | |||
|RECOMMENDED SEQUENCING || NextSeq500 - 75nt kit - 2-4 samples/flowcell | |||
|- | |||
|SUBMISSION || MIT - [https://mit.ilabsolutions.com/service_item/new/3381?spt_id=12903 ilabs] <BR> External contact biomicro@mit.edu | |||
|- | |||
|DELIVERY || FASTQ, SAM, BAM, Cell:Gene matrix | |||
|- | |||
|UNIT || PER ARRAY | |||
|} | |||
| | |||
The BioMicro Center collaborates closely with the Nanowell Core in the Koch Institute to provide support for the [https://shaleklab.com/resources/seq-well/ Seq-Well] protocol developed by Drs. Alex Shalek and Chris Love's groups. Seq-Well is based on the isolation of individual cells into microwells where cells are lysed and the associated mRNA is attached to polydT containing beads. <BR><BR> | |||
Seq-Well projects can be brought in two ways. For full service, please use the ilabs form linked on the left. At the end of the form it asks you to select several days on which the experiment can be done. The Nanowell Core will review the possible dates with you to fit it in the schedule. On that date, the core will confirm the quality of your cells prior to beginning the experiment. While 10,000 cells are typically used in a Seq-Well experiment, the core requests you plan to bring more cells - often counts from flow cytometers and other instruments are incorrect. Planning to bring more cells ensures you have the 10,000 desired. The BioMicro Center becomes involved in these projects at the exonuclease treatment step, where we take over the preparation of the library and quality control, as well as sequencing and analysis.<BR><BR> | |||
Many laboratories are also using reagents from the Nanowell core to begin doing Seq-Well on their own. For these labs, we strongly recommend at least confirming the quality of the cDNA on our Advanced Analytical before continuing through library preparation. The library preparation can also be done in the BioMicro Center. Please use the standard Illumina library preparation form and note that the samples are Seq-Well. We do have all the custom primers needed for preparation and the pricing is identical to other NexteraXT preps. | |||
|} | |||
--> | |||
Latest revision as of 13:43, 2 September 2025
HOME -- SEQUENCING -- LIBRARY PREP -- HIGH-THROUGHPUT -- COMPUTING -- OTHER TECHNOLOGY
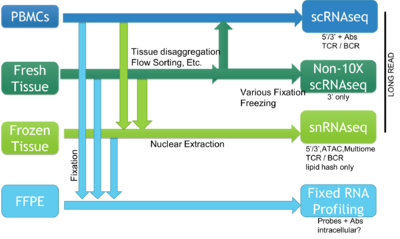
The BioMicro Center supports a broad range of methods for single cell sequencing. The choice of method depends heavily on the type of question being asked and the source material. The methods break down into those supporting individual cells characterized in single wells, methods that use droplet isolation and library preparation, and those that support rapid characterization of a modest number of transcripts using in situ sequencing (coming soon). These cell requirements for each method vary significantly so reviewing each method is valuable and we strongly encourage consultation with BMC staff prior to beginning the experiment.
.
10x CHROMIUM X[edit]
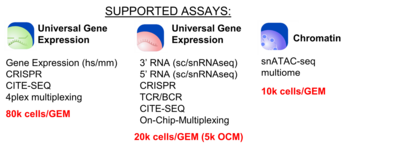
The BioMicro Center provides access to 10x Genomics library preparation as an assisted or a walk-up service. Added in 2023, The 10x Chromium X can handle a broad variety of methodologies now including 3', 5' and fixed RNA sequencing, ATAC and multiome Dropoff for assisted service is coordinated with Center staff with at least one week's lead time. Users should bring their single cell/nuclei suspension(s) in 1.5 mL microfuge tubes in the standard buffer at or around that time. Staff works with the user to intake the initial samples and proceed through the protocol with quality control checks at the appropriate steps.
Chromium X usage is also offered as a walkup service. Usage may be scheduled on the iLabs calendar after training by BMC staff. This still requires at least a week's lead time before usage. You will not have permission to schedule the equipment until you have been approved by BMC staff. Reagents are stocked in the BioMicro Center and we ask that you use our reagents. This allows us to get larger bulk discounts we can pass on to our users. Please note that we do include a fraction of the instrument usage cost in the cost of the chips and will charge this cost if you use your own chips.
The full 10x suite of software is installed on LURIA and is integrated into our analysis package.
scRNAseq / snRNAseq[edit]
CHROMATIN[edit]
|
10X Genomics[edit]
2) How many cells are captured in the Assay?
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
NAMOCELL SINGLE CELL SORTER[edit]
|
 The Namocell Single Cell Sorter allows users to sort cells into plates. The sorter uses microfluidics to sort single cells in 1uL of sheath fluid into a well. The instrument uses disposable cartridges to minimize contamination. The instrument integrates well with the TTP Labtech Mosquito HV which handles small reaction volumes. |
.
AVITI24 IN SITU SEQUENCING[edit]
*** coming in early 2026 ***
The AVITI24 enables direct in situ sequencing of samples, allowing for very rapid characterization of single cells. Cells can be grown or placed on the AVITI24 flowcell and processed. Runs are predicted to produce a few thousand reads plus cytostaining and antibody labeling, all within a day. More information will be released soon.

