BioMicroCenter:DNA HTL: Difference between revisions
| (4 intermediate revisions by 2 users not shown) | |||
| Line 111: | Line 111: | ||
| | | | ||
<br> | <br> | ||
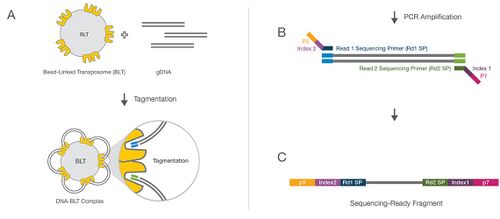
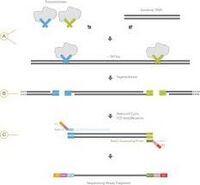
:Unlike XT, Nextera Flex utilizes bead-linked transposomes (BLT) to tagment and generate libraries from intact DNA samples. These BLTs control sizing during the tagmentation reaction by sterics, producing a much more consistent libraries with larger size distribution which makes it ideal for larger paired-end runs on sequencers. | :Unlike XT, Nextera Flex, also known as Illumina DNA Prep, utilizes bead-linked transposomes (BLT) to tagment and generate libraries from intact DNA samples. These BLTs control sizing during the tagmentation reaction by sterics, producing a much more consistent libraries with larger size distribution which makes it ideal for larger paired-end runs on sequencers. | ||
:Nextera Flex is currently run at a 1/10th miniaturization scale on the Mosquito HV with a broader dynamic range of input (1ng - 50ng) compared to XT. Nextera Flex self-normalizes at a certain input (if sample concentration is 3ng/µL or greater) and submissions do not have to be normalized so long as that threshold is met. Up to 384 libraries can be multiplexed using Nextera unique dual-indexes (UDI) which helps to avoid issues with 'barcode hopping'. | :Nextera Flex is currently run at a 1/10th miniaturization scale on the Mosquito HV with a broader dynamic range of input (1ng - 50ng) compared to XT. Nextera Flex self-normalizes at a certain input (if sample concentration is 3ng/µL or greater) and submissions do not have to be normalized so long as that threshold is met. Up to 384 libraries can be multiplexed using Nextera unique dual-indexes (UDI) which helps to avoid issues with 'barcode hopping'. | ||
::::[[IMAGE:Flex_Diagram.jpg | 500px]] | ::::[[IMAGE:Flex_Diagram.jpg | 500px]] | ||
| Line 219: | Line 219: | ||
| | | | ||
<br> | <br> | ||
[[image:16S_layout.jpg|thumb|200px| | [[image:16S_layout.jpg|thumb|200px|16S v4 region is amplified in standard BioMicro Center preps. Other regions can be substituted by submitting oligos with different variable regions. (Lopez-Aladid et al Sci. Rep. 2023)]] | ||
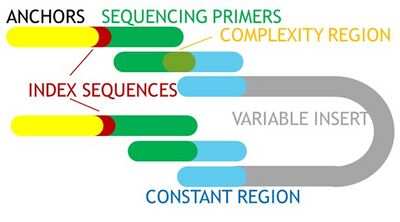
:The BioMicro Center offers 16S and amplicon sequencing for metagenomics projects. Derived from the [http://be.mit.edu/directory/eric-alm Alm Lab] with support from a pilot grant from [http://cehs.mit.edu/ MIT CEHS], our protocol uses a two step amplification to first expand the 16S population and add defined 3' and 5' sequences which are then used to add Illumina anchors and sequences. This two step method allows easy multiplexing and the ability change the amplicon insert sequence at minimal cost. | :The BioMicro Center offers 16S and amplicon sequencing for metagenomics projects. Derived from the [http://be.mit.edu/directory/eric-alm Alm Lab] with support from a pilot grant from [http://cehs.mit.edu/ MIT CEHS], our protocol uses a two step amplification to first expand the 16S population and add defined 3' and 5' sequences which are then used to add Illumina anchors and sequences. This two step method allows easy multiplexing and the ability change the amplicon insert sequence at minimal cost. | ||
[[image:16S_diagram.jpg|thumb|400px|Standard NGS amplicon design]] | |||
:The same method used for 16S is also applicable to other amplicons as well with minor adaption. Because we use a nested PCR, only the internal sequences need to be modified. The adapter sequences - Green:Blue - are the key element of this method. On the 5' end, they contain a "YRYR" sequence that introduces the complexity required for Illumina sequencing. | :The same method used for 16S is also applicable to other amplicons as well with minor adaption. Because we use a nested PCR, only the internal sequences need to be modified. The adapter sequences - Green:Blue - are the key element of this method. On the 5' end, they contain a "YRYR" sequence that introduces the complexity required for Illumina sequencing. | ||
<br> | <br> | ||
Latest revision as of 19:14, 9 June 2025
HOME -- SEQUENCING -- LIBRARY PREP -- HIGH-THROUGHPUT -- COMPUTING -- OTHER TECHNOLOGY
High-throughput DNA Library Preparation[edit]
The BioMicro Center offers high-throuhgput DNA library preparation (HTL-DNA) for both intact and fragmented samples as well as for metagenomics/amplicon generation. High-throughput library generation focuses on reducing price and processing time on a per sample basis. To do this, there are several key differences from our standard library preparation service. First, HTL-DNA services have set batch sizes (24 and 96) with fixed price points (i.e. charge for 16 samples will be the as 24 samples; similarly for 80 samples as 96). If under these batch sizes, DO MORE REPLICATES to simultaneously enhance experimental power and reduce price per sample. Some samples may fail and the cost of re-prepping those samples is NOT included in the quoted cost unless re-prep of the entire plate is deemed necessary. Re-preps can be done by hand, but at a higher rate. Second, the HTL option focuses on reducing price and processing time per sample, so some routine services provided with standard library preparation - such as initial and final quality control for all samples, sample arraying, or automatic re-prep of failed samples - are not built in. These services are available as add ons to the original service request.
Reminder: high throughput projects are treated with one condition (PCR cycles, fragmentation time, etc...). They are meant for allowing high replicates of the same samples. High throughput submissions for projects with multiple sub-projects from separate individuals combined on the same plate result in a low success rate due to the different conditions for each sub project. Therefore, htl submissions from multiple projects will tolerate and may expect, failure rates well over 10%.
FRAGMENTED DNA[edit]
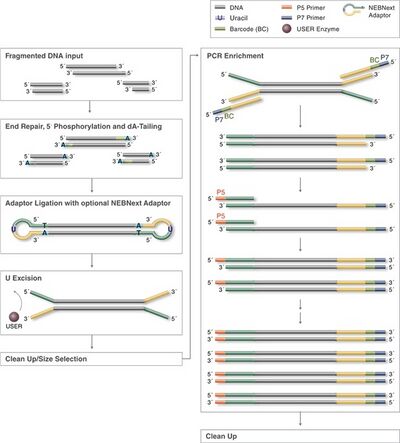
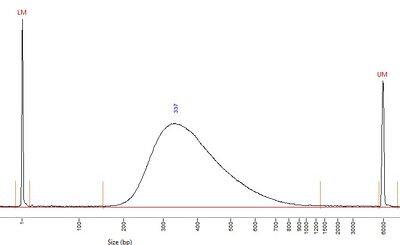
NEB Ultra II[edit]
|
|
|||||||||||||||||||||||||||||||||||||
INTACT GENOMES[edit]
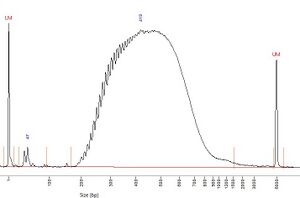
Nextera Flex[edit]
|
|
|||||||||||||||||||||||||||||||||||||
Nextera XT[edit]
|
|
|||||||||||||||||||||||||||||||||||||
16S / AMPLICON SEQUENCING[edit]
|
|
|||||||||||||||||||||||||||||||||||||||||||
USEFUL INFORMATION[edit]
DNA Pre-load Submissions[edit]
- Describes samples that upon submission to the BMC can be immediately plugged into the preparatory method that has been specified in the project submission form. This option is provided to reduce library preparation costs and expedite sample processing by minimizing hands-on time by a BMC staff. The specific requirements are listed for each preparatory method (excluding 16S) in the tables above. These specifications must be met in order to qualify as a pre-load, submissions otherwise may be subject to additional charges.
- Due to the nature of a pre-load submission, the entire volume of sample submitted will be used and as such, additional services are not typically offered unless coordinated with a BMC technician. It is important to keep this in mind when submitting precious samples and the BMC recommends to save a portion of sample when possibly.
Quadrant Layout[edit]
- At the BioMicro Center, we organize our high-throughput projects in 384-well plates using a quadrant layout. With quadrant 1 (Q1) representing one 96-well plate, quadrant 2 (Q2) representing a second 96-well plate and so on up to Q4. We ask that plates being submitted start with Q1 and arrayed column-wise. Below is a diagram illustrating the desired filled quadrant.
Quadrant 1 of 384-well plate 
|
Normalization[edit]
- Normalization is referring to bringing all samples to a uniform concentration. For library preparation, uniform input mass is necessary for proper library generation and as such, calculations for normalization should be based upon concentration in ng/µL. This is in contrast to normalization for pooling of final libraries, where calculations are based upon concentration in nM since the number of molecules is important to achieve a desired read distribution when sequencing.
Unique versus Combinatorial Dual Indexing[edit]
- Combinatorial dual indexing is a technique that uses a set of Index 1 (denoted as i7) and Index 2(denoted as i5) barcodes that are combined in a manner such that it produces distinct i7 and i5 pairs which increases multiplexing capacity for sequencing. With combinatorial dual indexes, each i7 and i5 is shared among other samples on the same plate, typically with i5's repeating across rows and i7's down columns. These combinations are unique but individual indexes used are not. However, a phenomenon known as 'index hopping' has been observed when sequencing multiplexed libraries that followed single or combinatorial indexing schemes with newer Illumina platforms utilizing ExAmp chemistry such as the NovaSeq (Costello et al., 2018). This swapping of indexes causes reads to be mis-assigned and subsequently excluded from further analysis. The primary strategy employed to mitigate the effects of index hopping is through the utilization of unique dual indexes.
- Unique dual indexes (UDI) are non-redundant indexes where each i5 and i7 has a distinct index sequence. As opposed to combinatorial dual indexing, an i7 and i5 index is never repeated nor shared among other samples (i.e. for a 96-well UDI index plate, there are 96 unique i7's and 96 unique i5's). Both the combinations and individual indexes used are unique, and as a result the frequency of mis-assigned reads due to index hopping is greatly reduced. The BioMicro Center recommends that UDI's always be used when sequencing on the NovaSeq. The number of libraries that can be multiplexed and sequenced on a single lane is determined by the total number of UDI's provided for each library preparation method.