BioMicroCenter:Oxford Nanopore Technologies: Difference between revisions
| (12 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{BioMicroCenter}} | {{BioMicroCenter}} | ||
<br><br> | <br><br> | ||
The MIT BioMicro Center has | The MIT BioMicro Center has two PromethION P2 Solos, each capable of running two concurrent flowcells. The Center supports all current standard ONT library prep protocols including both ligation and tagmentation methods as well as direct RNA measurement. Run protocols such as adaptive sampling are available. Each flowcell is run to completion and not reused - prices are for full flowcells. | ||
<br><br> | <br><br> | ||
==Oxford Nanopore Sequencing== | ==Oxford Nanopore Sequencing== | ||
| Line 24: | Line 24: | ||
* Quality Control | * Quality Control | ||
* [[BioMicroCenter:NanoPore_Library_Prep|Nanopore library preparation]] | * [[BioMicroCenter:NanoPore_Library_Prep|Nanopore library preparation]] | ||
* Modified basecalling available on request | |||
* Reference FASTA genome and BED regions file required for adaptive sampling | |||
|- | |- | ||
|DATA FORMATS | |DATA FORMATS | ||
| Line 40: | Line 42: | ||
|} | |} | ||
| | | | ||
A variety of preparation kits allow DNA | A variety of preparation kits allow DNA or direct RNA to be sequenced. The input amount necessary for each type of sample varies according to the desired result. For the longest reads of genomic DNA, micrograms worth of clean starting material provide the highest quality of data, but not the highest amounts of reads. | ||
<br><br> | <br><br> | ||
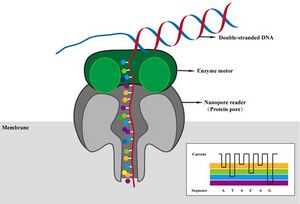
Nanopore library contains adaptor with both motor protein and tether. If tether interacts with a nanopore successfully, the motor protein will shift the molecule into and through the protein nanopore. The voltage set across the nanopore shifts as base pairs move through the pore. Nanopores are arranged in an array such that multiple molecules can be sequenced simultaneously. Sequencing and demultiplexing all occur in real time providing fast5 and/or fastq. | |||
<br><br> | <br><br> | ||
Contaminants known to cause issues for the nanopores | Contaminants known to cause issues for the nanopores even at low amounts include: | ||
*EDTA | |||
*Alcohols (EtOH, iPrOH | |||
*High concentrations of salts (NaCl, guanidinium chloride,guanidinium isothiocyanate | |||
*phenol | |||
<br><br> | |||
Modified basecalling is available on request. Not all desired basecalled modifications are possible in a single sequencing run with the current chemistries and basecalling software, but trace data (fast5) can be taken and re-basecalled using different basecallers. Re-basecalling can be done as a separate bioinformatic project. | |||
<br><br> | |||
For adaptive sampling, the user must provide at least a FASTA genome file and, optionally a BED file with the region(s) of interest. It is important that for both enrichment and depletion such region(s) only entail 0.1-10% of expected reads | |||
| | | | ||
[[Image:Nanopore.jpg|300px|middle]] | [[Image:Nanopore.jpg|300px|middle|Ji, CM et al (2024). Viruses, 16(5), 798.]] <br><br> | ||
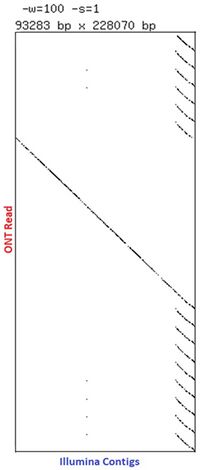
[[Image:Spanreads.jpg|200px|middle|Page Lab assembled ultra long read gDNA data showing a long read spanning Illumina contigs]] <br><br> | |||
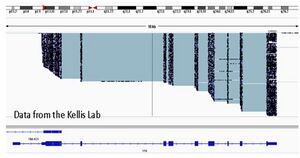
[[Image:IGV_vimentin.jpg|300px|middle|IGV spanning the Vimentin gene with assembled amplified cDNA]] | |||
|} | |} | ||
==Oxford Nanopore | ==Oxford Nanopore Sequencer== | ||
{| class="wikitable" border=1 | {| class="wikitable" border=1 | ||
!width=100| SPEC | !width=100| SPEC | ||
| Line 57: | Line 69: | ||
|[[Image:PromethION.jpg|center|200px]]© Oxford Nanopore Technologies | |[[Image:PromethION.jpg|center|200px]]© Oxford Nanopore Technologies | ||
|- | |- | ||
| '''AVG | | '''AVG OUTPUT/FLOWCELL'''<BR> | ||
| | | 50 Gb (highly variable) | ||
50 Gb | |||
|- | |- | ||
|'''MAX TIME/FLOWCELL''' | |'''MAX TIME/FLOWCELL''' | ||
| | | 3 days (adjustable) | ||
3 days (adjustable) | |||
|} | |} | ||
Latest revision as of 12:29, 18 July 2025
HOME -- SEQUENCING -- LIBRARY PREP -- HIGH-THROUGHPUT -- COMPUTING -- OTHER TECHNOLOGY
The MIT BioMicro Center has two PromethION P2 Solos, each capable of running two concurrent flowcells. The Center supports all current standard ONT library prep protocols including both ligation and tagmentation methods as well as direct RNA measurement. Run protocols such as adaptive sampling are available. Each flowcell is run to completion and not reused - prices are for full flowcells.
Oxford Nanopore Sequencing[edit]
|
A variety of preparation kits allow DNA or direct RNA to be sequenced. The input amount necessary for each type of sample varies according to the desired result. For the longest reads of genomic DNA, micrograms worth of clean starting material provide the highest quality of data, but not the highest amounts of reads.
|
Oxford Nanopore Sequencer[edit]
| SPEC | PromethiON P2 Solo |
|---|---|
| SEQUENCER |  |
| AVG OUTPUT/FLOWCELL |
50 Gb (highly variable) |
| MAX TIME/FLOWCELL | 3 days (adjustable) |