BioMicroCenter:Singular Sequencing: Difference between revisions
No edit summary |
|||
| (57 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
== Singular Sequencing == | == Singular Sequencing == | ||
The Center currently hosts a G4 platform by [https://singulargenomics.com/| Singular Genomics]. Singular’s G4 platform is a short-read mid-throughput sequencing platform delivering adequate sequencing data through its proprietary sequencing-by-synthesis chemistry at a significantly competitive price point. | The Center currently hosts a G4 platform by [https://singulargenomics.com/| Singular Genomics]. Singular’s G4 platform is a short-read mid-throughput sequencing platform delivering adequate sequencing data through its proprietary sequencing-by-synthesis chemistry at a significantly competitive price point. | ||
=== | {| class="wikitable" border=1 | ||
!width=100| Service | |||
!width=250| Singular Sequencing | |||
|- | |||
|INPUT || Singular libraries | |||
|- | |||
|MIN VOLUME || 12 uL | |||
|- | |||
< | |MIN CONCENTRATION || 4 nM | ||
|- | |||
|INCLUDED SERVICES | |||
| | |||
* Quality Control:Fragment Analyzer and qPCR | |||
* Singular Sequencing | |||
* Demultiplexing | |||
|- | |||
|ADDITIONAL SERVICES || | |||
* Quality Control | |||
* Singular library preparation | |||
|- | |||
|DATA FORMATS | |||
| | |||
*FASTQ (stored 1 year) | |||
|- | |||
|QUALITY CONTROL | |||
| | |||
* [https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ FASTQC] | |||
* Basic run metrics (alignment rate, complexity) | |||
* Basic RNAseq metrics (where applicable) | |||
* Basic paired end metrics (where applicable) | |||
* Contamination checks | |||
|- | |||
|PRICING || [[BioMicroCenter:Pricing#SINGULAR_G4|LINK]] | |||
|- | |||
|SUBMISSION | |||
| | |||
* MIT - [https://mit.ilabsolutions.com/service_item/new/3381?spt_id=3863 ilabs] | |||
|- | |||
|} | |||
Compared to a typical Illumina library, a Singular index 1 would be the reverse compliment of the i5 index, whereas index 2 would be the reverse compliment of the i7 index. | |||
===Lane-by-lane Requirements=== | |||
*Must be 50 or 150 paired-end dual indexed | |||
*Pool must be compatible with TruSeq, Nextera, TruSeq, smRNA and Solexa sequencing primers | |||
Minimum reads per lane are guaranteed if: | |||
*the BMC has performed quality control, | |||
*the samples are high-complexity, especially for the first 10 nt of read 1, and | |||
*submitted libraries are at least 2 nM. | |||
All other requests, including use of custom sequencing primers require a full flowcell. Illumina libraries which have been converted in the Center will also require a full flowcell. | |||
*Min custom primer submission: 20 uL at 100 uM | |||
[[IMAGE:IndexhopF3.jpeg|left|250px|Index hopping for a standard set of Singular libraries]] [[IMAGE:PPPPF3.jpeg|right|200px|Percent Perfect Plot for Singular libraries 150PE]] | |||
Index hopping on Singular runs has been low. The performance of an F3 flowcell mimics other short-read 4 color chemistry sequencers with patterned flowcells. | |||
<br><br><br><br> | |||
== The G4 Platform == | == The G4 Platform == | ||
| Line 23: | Line 70: | ||
|'''SEQUENCER'''||[[Image:2023_Singular_G4.jpg|center|200px]] | |'''SEQUENCER'''||[[Image:2023_Singular_G4.jpg|center|200px]] | ||
|- | |- | ||
| '''READS/LANE'''<BR> Low number is minimum per lane for standard | | '''READS/LANE'''<BR> Low number is minimum per lane for standard libraries|| | ||
* F3 per Lane: 50-100 M | * F3 per Lane: 50-100 M | ||
|- | |- | ||
|'''RUN TIME'''|| | |'''RUN TIME'''|| | ||
Latest revision as of 17:26, 9 June 2025
Singular Sequencing[edit]
The Center currently hosts a G4 platform by Singular Genomics. Singular’s G4 platform is a short-read mid-throughput sequencing platform delivering adequate sequencing data through its proprietary sequencing-by-synthesis chemistry at a significantly competitive price point.
| Service | Singular Sequencing |
|---|---|
| INPUT | Singular libraries |
| MIN VOLUME | 12 uL |
| MIN CONCENTRATION | 4 nM |
| INCLUDED SERVICES |
|
| ADDITIONAL SERVICES |
|
| DATA FORMATS |
|
| QUALITY CONTROL |
|
| PRICING | LINK |
| SUBMISSION |
|
Compared to a typical Illumina library, a Singular index 1 would be the reverse compliment of the i5 index, whereas index 2 would be the reverse compliment of the i7 index.
Lane-by-lane Requirements[edit]
- Must be 50 or 150 paired-end dual indexed
- Pool must be compatible with TruSeq, Nextera, TruSeq, smRNA and Solexa sequencing primers
Minimum reads per lane are guaranteed if:
- the BMC has performed quality control,
- the samples are high-complexity, especially for the first 10 nt of read 1, and
- submitted libraries are at least 2 nM.
All other requests, including use of custom sequencing primers require a full flowcell. Illumina libraries which have been converted in the Center will also require a full flowcell.
- Min custom primer submission: 20 uL at 100 uM

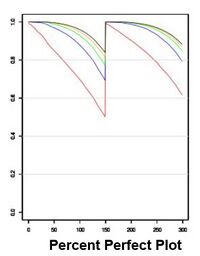
Index hopping on Singular runs has been low. The performance of an F3 flowcell mimics other short-read 4 color chemistry sequencers with patterned flowcells.

