BioMicroCenter:RNA LIB: Difference between revisions
| (10 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{BioMicroCenter}} | {{BioMicroCenter}} | ||
[[Image:RNA-Seq_pres.jpg|thumb|300px|right|Wang | [[Image:RNA-Seq_pres.jpg|thumb|300px|right|Wang Z, et al. Nat Rev Genet 2009]] | ||
The BioMicro Center supports a broad variety of standard library preparation methods for RNAseq. The choice of method is highly dependent on the type of input, the amount of input RNA available, and the quality of the input RNA. The key in all RNAseq methods is the avoidance of ribosomal RNA, which would dominate the library preparation. Below is a summary of the methods we utilize routinely in the core. For High-Throughput RNA library preparation, please see our page for [[BioMicroCenter:RNA_HTL|methods designed specifically for large sample batches.]] | The BioMicro Center supports a broad variety of standard library preparation methods for RNAseq. The choice of method is highly dependent on the type of input, the amount of input RNA available, and the quality of the input RNA. The key in all RNAseq methods is the avoidance of ribosomal RNA, which would dominate the library preparation. Below is a summary of the methods we utilize routinely in the core. For High-Throughput RNA library preparation, please see our page for [[BioMicroCenter:RNA_HTL|methods designed specifically for large sample batches.]] | ||
| Line 9: | Line 9: | ||
! Method Recommended | ! Method Recommended | ||
|- | |- | ||
| >10ng || RIN:>8.0 || [[BioMicroCenter:RNA_LIB#NEB_Ultra_II_Directional_RNA_with_Poly(A)_Selection|NEB Poly A | | >10ng || RIN:>8.0 || [[BioMicroCenter:RNA_LIB#NEB_Ultra_II_Directional_RNA_with_Poly(A)_Selection|NEB Poly A]] | ||
|- | |- | ||
| >50ng || DV200>0.2 || | | >50ng || DV200>0.2 || NEB ribosomal depletion followed by NEB UltraII prep. | ||
|- | |- | ||
| 10pg-25ng || RIN:9.0 || [[BioMicroCenter:RNA_LIB#Clontech_SMARTseq_Low-Input|Clontech SMARTer v4]] | | 10pg-25ng || RIN:9.0 || [[BioMicroCenter:RNA_LIB#Clontech_SMARTseq_Low-Input|Clontech SMARTer v4]] | ||
| Line 75: | Line 75: | ||
|} | |} | ||
<!-- commenting Kapa out | |||
==[https://sequencing.roche.com/en-us/products-solutions/by-category/library-preparation/rna-library-preparation/kapa-mrna-hyperprep-kits.html Kapa mRNA Hyperprep] == | ==[https://sequencing.roche.com/en-us/products-solutions/by-category/library-preparation/rna-library-preparation/kapa-mrna-hyperprep-kits.html Kapa mRNA Hyperprep] == | ||
{| | {| | ||
| Line 96: | Line 97: | ||
|} | |} | ||
| | | | ||
This workflow is very similar to Illumina's TruSeq chemistry at lower cost and is streamlined for automation. This chemistry uses polyT beads to isolate the mRNA from the rRNA and tRNA. The use of these beads requires that the RNA be of very high quality or only the 3' end of transcripts will be isolated. Purified mRNA is then fragmented with metal and random priming is used to convert the sample to cDNA. Once double-stranded cDNA is generated, LMPCR is performed to create the indexed Illumina library. The BioMicro Center offers mRNA HyperPrep as a single sample reaction or in [[BioMicroCenter:RNA_HTL|batches of 24 done on the TecanEvo 150s.]] | |||
| | | | ||
|} | |} | ||
| Line 131: | Line 128: | ||
| | | | ||
|} | |} | ||
end of kapa --> | |||
== [http://www.clontech.com/US/Products/cDNA_Synthesis_and_Library_Construction/cDNA_Synthesis_Kits/Ultra_Low_Input_RNA_cDNA_Synthesis Clontech SMARTseq Low-Input] == | == [http://www.clontech.com/US/Products/cDNA_Synthesis_and_Library_Construction/cDNA_Synthesis_Kits/Ultra_Low_Input_RNA_cDNA_Synthesis Clontech SMARTseq Low-Input] == | ||
{| | {| | ||
| Line 149: | Line 146: | ||
|} | |} | ||
| | | | ||
For samples with less then 50ng of input, the BioMicro Center utilizes the [http://www.clontech.com/US/Products/cDNA_Synthesis_and_Library_Construction/cDNA_Synthesis_Kits/Ultra_Low_Input_RNA_cDNA_Synthesis Clontech SMARTseq v4 system]. This system differs from the TruSeq chemistry in that it begins with cDNA generation using polyT priming followed by strand switching oligos. The use of polyT priming requires the RNA to be of high quality. Full length double-stranded cDNAs are generated and amplified by PCR. These cDNAs | For samples with less then 50ng of input, the BioMicro Center utilizes the [http://www.clontech.com/US/Products/cDNA_Synthesis_and_Library_Construction/cDNA_Synthesis_Kits/Ultra_Low_Input_RNA_cDNA_Synthesis Clontech SMARTseq v4 system]. This system differs from the TruSeq chemistry in that it begins with cDNA generation using polyT priming followed by strand switching oligos. The use of polyT priming requires the RNA to be of high quality. Full length double-stranded cDNAs are generated and amplified by PCR. These cDNAs can be prepared into NGS short-read libraries using the NexteraXT chemistry from Illumina. Data from this system is of similar quality to samples created with Illumina TruSeq chemistry but is not stranded. Single samples can be prepared by hand. [[BioMicroCenter:RNA_HTL|Batches of 24, 96 or 384 samples]] can be prepared using the older SMARTseq v2 chemistry on the Mosquito HV resulting in significantly lower costs/sample. | ||
|} | |} | ||
| Line 171: | Line 167: | ||
|} | |} | ||
| | | | ||
For samples with less then 100ng of input and restricted input amounts, our kit of choice is the Clontech SMARTer Stranded Total RNAseq Kit - Pico Input -- or more simply, Clontech ZapR . This kit utilizes the same template switching as the v4 kit but uses random primers on fragmented RNA. The key is the ZapR enzyme which is used post library production to, in a targeted manner, cause breaks in Illumina library molecules that contain rRNA reads. These breaks make the rRNA containing molecules unreadable. | For samples with less then 100ng of input and restricted input amounts, our kit of choice is the Clontech SMARTer Stranded Total RNAseq Kit - Pico Input -- or more simply, Clontech ZapR . This kit utilizes the same template switching as the v4 kit but uses random primers on fragmented RNA. The key is the ZapR enzyme which is used post library production to, in a targeted manner, cause breaks in Illumina library molecules that contain rRNA reads. These breaks make the rRNA containing molecules unreadable. This chemistry can be utilized in HTL format as well. <BR><BR> | ||
In analyzing data from this kit, we have observed that the first few nucleotides from many reads appear to have a very high mismatch rate, particularly from low input samples or samples that possibly may not be as clean as desired. We believe this is a results of the template switching and random priming. A 5nt trim from the 5'end of the read can significantly improve data quality. | In analyzing data from this kit, we have observed that the first few nucleotides from many reads appear to have a very high mismatch rate, particularly from low input samples or samples that possibly may not be as clean as desired. We believe this is a results of the template switching and random priming. A 5nt trim from the 5'end of the read can significantly improve data quality. | ||
| | | | ||
|} | |} | ||
== Additional Chemistries Available in the BioMicro Center == | == Additional Chemistries Available in the BioMicro Center == | ||
=== [[BioMicroCenter:PippinPrep|Size Selection]] === | === [[BioMicroCenter:PippinPrep|Size Selection]] === | ||
For some applications of RNAseq, such as splice choice determination, having a precise knowledge of the insert size is critical. While | For some applications of RNAseq, such as splice choice determination, having a precise knowledge of the insert size is critical. While a standard SPRI clean does provide some size selection (typically restricting fragments to between 150 and 350bp), this can be too wide for some methodologies. In these cases, after libraries are amplified, they can be run on the [[BioMicroCenter:PippinPrep|Sage BluePippin]] (either singly or pooled). Here the size distribution can be much tighter, with most of the DNA fragments being within a 50nt range. | ||
Latest revision as of 14:52, 2 May 2025
HOME -- SEQUENCING -- LIBRARY PREP -- HIGH-THROUGHPUT -- COMPUTING -- OTHER TECHNOLOGY
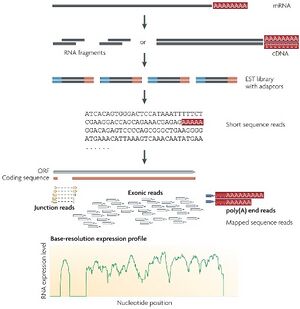
The BioMicro Center supports a broad variety of standard library preparation methods for RNAseq. The choice of method is highly dependent on the type of input, the amount of input RNA available, and the quality of the input RNA. The key in all RNAseq methods is the avoidance of ribosomal RNA, which would dominate the library preparation. Below is a summary of the methods we utilize routinely in the core. For High-Throughput RNA library preparation, please see our page for methods designed specifically for large sample batches.
| Amount of total RNA | Quality of RNA | Method Recommended |
|---|---|---|
| >10ng | RIN:>8.0 | NEB Poly A |
| >50ng | DV200>0.2 | NEB ribosomal depletion followed by NEB UltraII prep. |
| 10pg-25ng | RIN:9.0 | Clontech SMARTer v4 |
| 1ng-1ug | DV200>0.2 | Clontech Pico Ribosomal Depletion (ZapR). |
| smallRNA | NA | Qiagen miRNA kit or Clontech microRNA. |
NEB Ultra II Directional RNA with Poly(A) Selection[edit]
|
The BioMicro Center utilizes the NEBNext Ultra II Directional RNA preparation with poly(A) selection kit to prepare standard RNA libraries. The selection of mRNA transcripts occurs by hybridization of the poly(A)-tail to magnetic oligo-d(T) beads. We can reliably generate RNA-Seq libraries with inputs ranging from 10ng to 100ng (recommended input of > 50ng). High-quality samples are required because the method is reliant on poly(A)-tails during RNA isolation. We also offer a high-throughput protocol. |
NEB Ultra II Directional RNA with rRNA Depletion (H/M/R)[edit]
|
NEBNext Ultra II Directional RNA preparation with ribosomal RNA (rRNA) depletion is used to deplete rRNA by enzymatic degradation using single-stranded DNA probes that target rRNAs, leaving all other RNA species present and available for library preparation. This depletion method is agnostic to input sample quality and is the go-to method if samples don't meet quality requirements for poly(A) selection. This method is suited for inputs ranging from 5ng to 100ng (recommended input of > 50ng). Due to probe design used in the depletion, this method is currently only available for Human/Mouse/Rat samples. We also offer a high-throughput protocol. |
Clontech SMARTseq Low-Input[edit]
|
For samples with less then 50ng of input, the BioMicro Center utilizes the Clontech SMARTseq v4 system. This system differs from the TruSeq chemistry in that it begins with cDNA generation using polyT priming followed by strand switching oligos. The use of polyT priming requires the RNA to be of high quality. Full length double-stranded cDNAs are generated and amplified by PCR. These cDNAs can be prepared into NGS short-read libraries using the NexteraXT chemistry from Illumina. Data from this system is of similar quality to samples created with Illumina TruSeq chemistry but is not stranded. Single samples can be prepared by hand. Batches of 24, 96 or 384 samples can be prepared using the older SMARTseq v2 chemistry on the Mosquito HV resulting in significantly lower costs/sample. |
Clontech SMARTer Stranded Total RNA-Seq Kit - Pico Input Mammalian -- aka Clontech ZapR[edit]
|
For samples with less then 100ng of input and restricted input amounts, our kit of choice is the Clontech SMARTer Stranded Total RNAseq Kit - Pico Input -- or more simply, Clontech ZapR . This kit utilizes the same template switching as the v4 kit but uses random primers on fragmented RNA. The key is the ZapR enzyme which is used post library production to, in a targeted manner, cause breaks in Illumina library molecules that contain rRNA reads. These breaks make the rRNA containing molecules unreadable. This chemistry can be utilized in HTL format as well. In analyzing data from this kit, we have observed that the first few nucleotides from many reads appear to have a very high mismatch rate, particularly from low input samples or samples that possibly may not be as clean as desired. We believe this is a results of the template switching and random priming. A 5nt trim from the 5'end of the read can significantly improve data quality. |
Additional Chemistries Available in the BioMicro Center[edit]
Size Selection[edit]
For some applications of RNAseq, such as splice choice determination, having a precise knowledge of the insert size is critical. While a standard SPRI clean does provide some size selection (typically restricting fragments to between 150 and 350bp), this can be too wide for some methodologies. In these cases, after libraries are amplified, they can be run on the Sage BluePippin (either singly or pooled). Here the size distribution can be much tighter, with most of the DNA fragments being within a 50nt range.

